Comparing common compression tools using real-world data¶
Compression tools can significantly reduce the amount of disk space consumed by your data. In this article, we will look at the effectiveness of some compression tools on real-world data sets, make some recommendations, and perhaps persuade you that compression is worth the effort.
Compression on a single processor core¶
Lossless compression of files is a great way to save space, and therefore money, on storage costs. Not all compression tools are equal and your experience will vary depending of which of the wide range of available compression tools you use. There is also a historical perception that compression and decompression are slow and time consuming, introducing unnecessary delays into the workflow.
Although you will need to decompress any compressed data before it is used, decompression can be very rapid and most of the time won't introduce significant delay to your workflow.
We will confine all of the tools to use a single core, to enable direct comparison.
Tools¶
We will be performing tests using the following popular open-source tools:
-
gzip v1.10 (compiled from source)
-
xz v5.2.2, (CentOS repository)
-
bzip2 v1.0.6 (CentOS repository)
-
pigz v2.4 -a parallel implementation of gzip (compiled from source)
-
zstd v1.4.1 -touted as a fast compression algorithm (compiled from source)
Other tools considered include lrzip and lz4.
Some of these tools offer multi-threaded options, which have stunning results
in conjunction with the QMUL HPC Cluster, where typically the Research Data
resides.
Datasets¶
Two datasets were selected for testing:
- a human genome GRCh38 reference assembly file, size ~3.3GB uncompressed
- the Linux kernel v5.2, size 831MB uncompressed
The Human Genome reference file only contains the characters {g,a,c,t,G,A,C,T,N} but is an example of a genome reference file commonly used in bioinformatics. Effective compression for bioinformatics is important due to frequent use of large data files. The scope of this article is to investigate the efficacy of a range of generalist tools to work with a variety of datasets, while bearing in mind that other specialist tools may produce better compression for a narrow range of data types. For example, GeCo compressed the human genome file to around half the size of the general tools. However, these specialist tools carry risk; some of the projects are abandoned, and produce proprietary formats, and don't necessarily decompress to a file identical to the original! It's better to stick to popular open-source tools under active development.
The Linux kernel has over 20 million lines of code, mostly heavily commented C language.
The files were obtained via the following Linux commands, and subsequently uncompressed onto high-performance networked NVME storage.
wget ftp://ftp.ncbi.nlm.nih.gov/genomes/all/GCF/000/001/405/GCF_000001405.39_GRCh38.p13/GCF_000001405.39_GRCh38.p13_genomic.fna.gz
wget https://cdn.kernel.org/pub/linux/kernel/v5.x/linux-5.2.tar.xz
Method¶
Compression tests were run on a QMUL HPC Cluster
node with Intel Gold 6126 Skylake
Processors running CentOS 7.6. Single-core tests were run while the machine was
idle, to avoid any thermal throttling from other computations running on the
machine. For each compression tool, the test data was compressed once for each
available compression level, observing runtime and resulting file sizes using
time and ls commands. The runs were repeated to confirm results.
pigz and zstd were constrained to use one thread for the single-core tests,
since they attempt to use more cores by default.
Definitions¶
The compression speed is defined as: (uncompressed data size in GB)/(time taken to compress in seconds)
Decompression speed is defined as : (uncompressed data size in GB)/(time taken to decompress in seconds)
Single-Core Test Results¶
Compiler optimisation¶
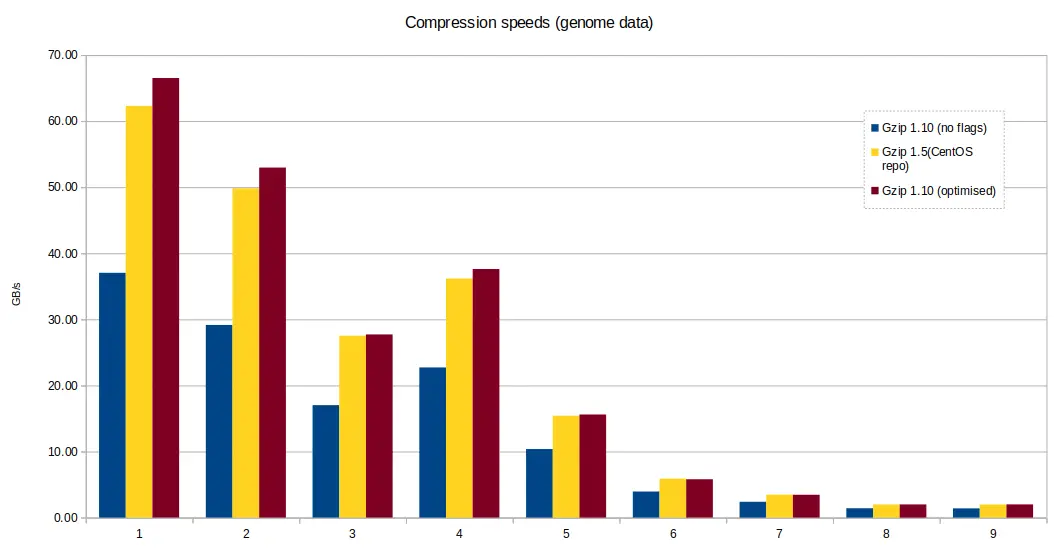
Some preliminary tests were run to compare gzip binaries compiled with and
without gcc compiler optimisation -O3 -march=native. There was on average, a
10-20% performance loss when compiling gzip 1.10 from source with gcc,
without any compiler flags, versus gzip 1.5 available in the CentOS
repository, and gzip 1.10 with -O3 -march=native flags.
The CentOS rpm package had been compiled with the following standard CentOS toolchain flags:
-O2 -g -pipe -Wall -Wp,-D_FORTIFY_SOURCE=2 -fexceptions -fstack-protector-strong \
--param=ssp-buffer-size=4 -grecord-gcc-switches -m64 -mtune=generic
For the following tests, we continued with gzip 1.10 compiled with the gcc
optimisation.
Genome dataset¶
Speeds¶
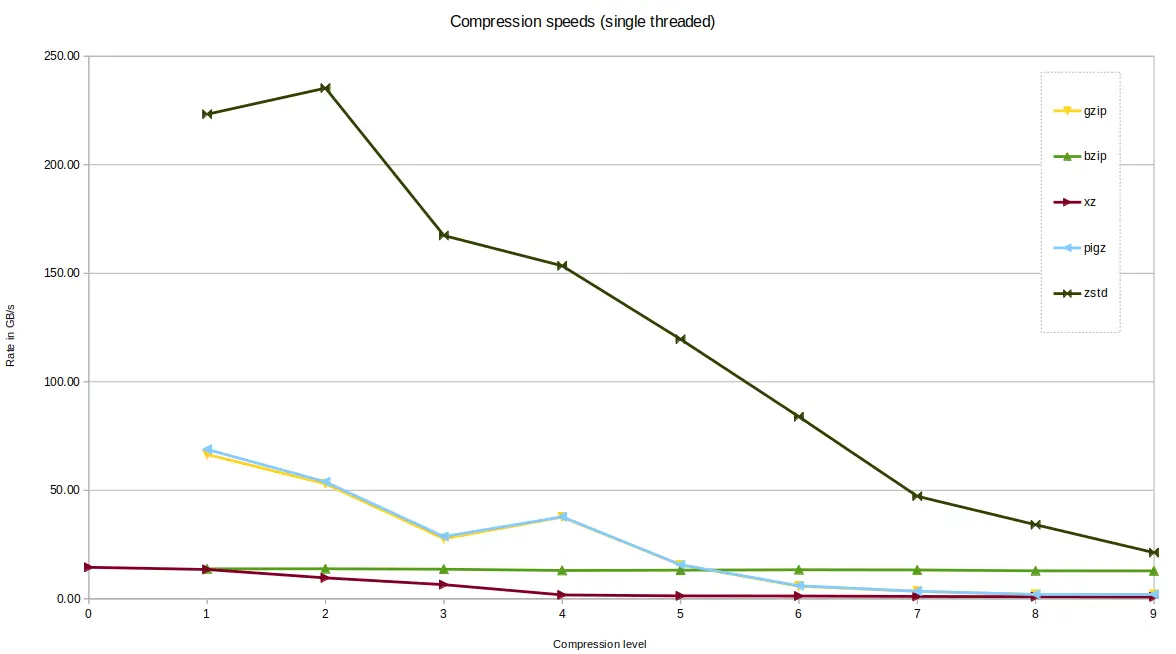
zstd was fastest of the tools tested. zstd in fact offers 19 compression
levels, the first 9 are shown on this graph only.
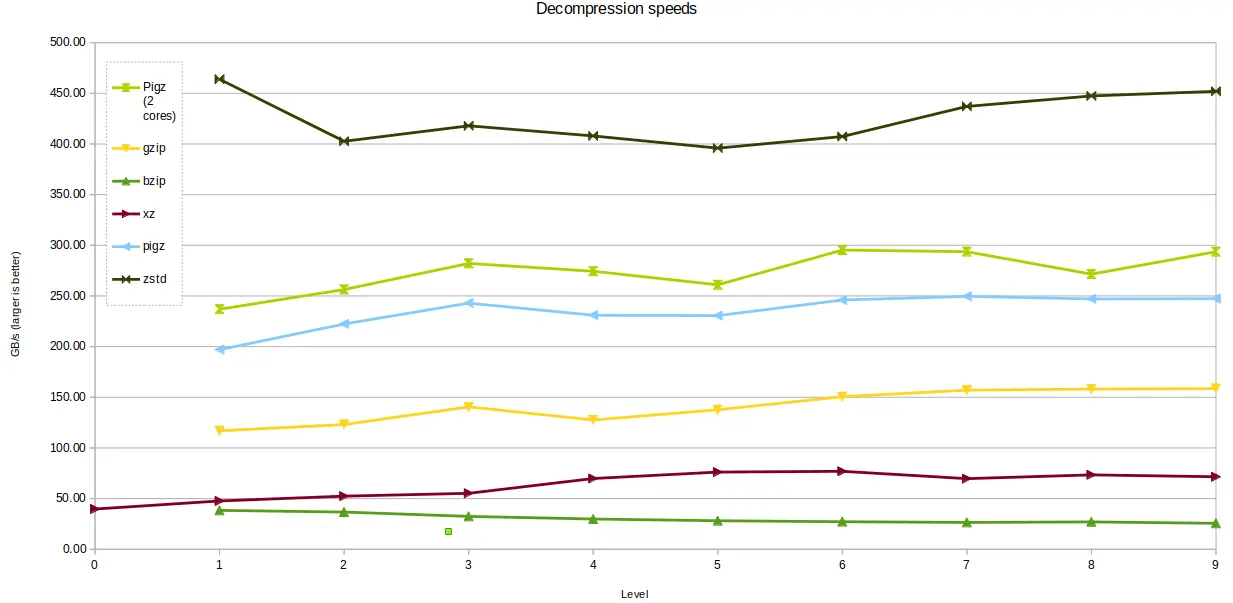
Most decompression is single-threaded, except for pigz which can offer
improved performance if allowed to use two cores. Both single-core and dual-core
pigz results are shown in the chart below. zstd was the clear winner for
decompression speed, offering around ten times the performance of bzip2 and
xz
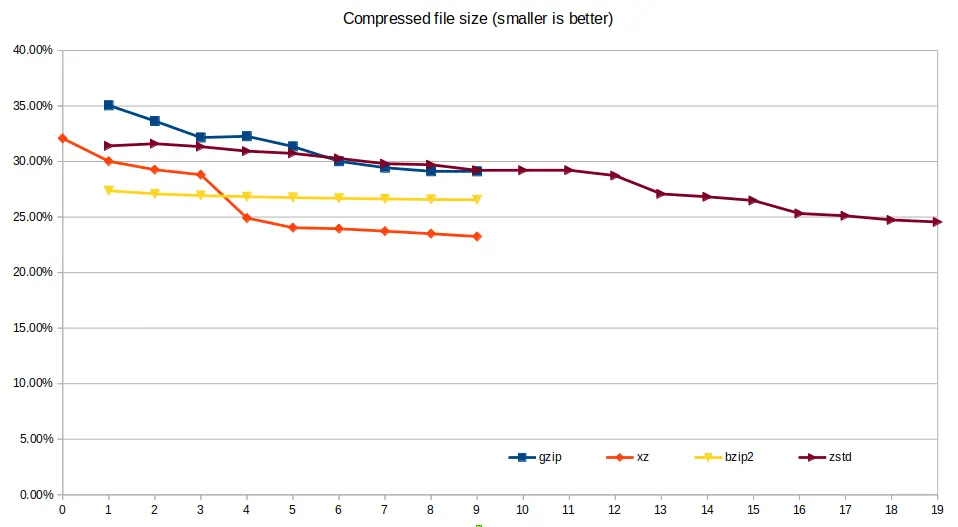
File size¶
xz was able to deliver the smallest file size for the compressed genomics
data, however at a cost of speed. gzip and pigz both result in the same
file size and are only represented once in the below chart; the only difference
is the computational performance.
Linux kernel dataset¶
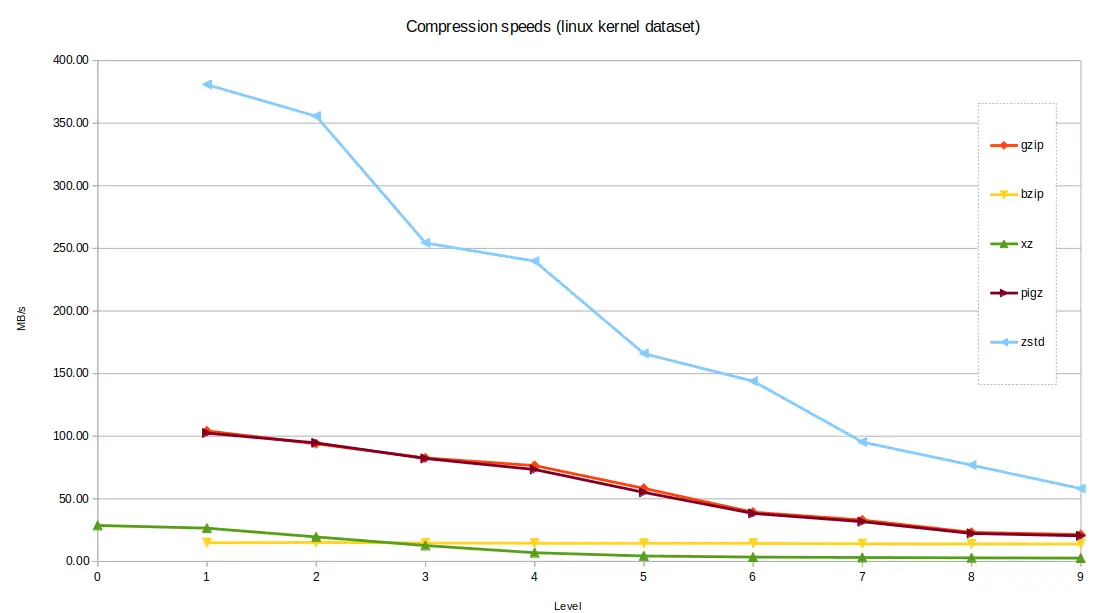
Processing speeds¶
The Linux kernel dataset contains a lot of C Code and English text. It serves well as an example of a large code base, and should compress and decompress better than the genome file.
zstd performed very well again, compared with the alternatives, with bzip2
and xz the slowest. When considering the fastest option for each tool, at
level 1 compression, zstd was able to produce a file 20.5% of the original
size in a blistering 2.18 seconds. At the other end of the scale, bzip2 took
56 seconds on its fastest setting, producing 17.5% compression. The fastest
setting of xz took 29 seconds and produced around 18% compression.
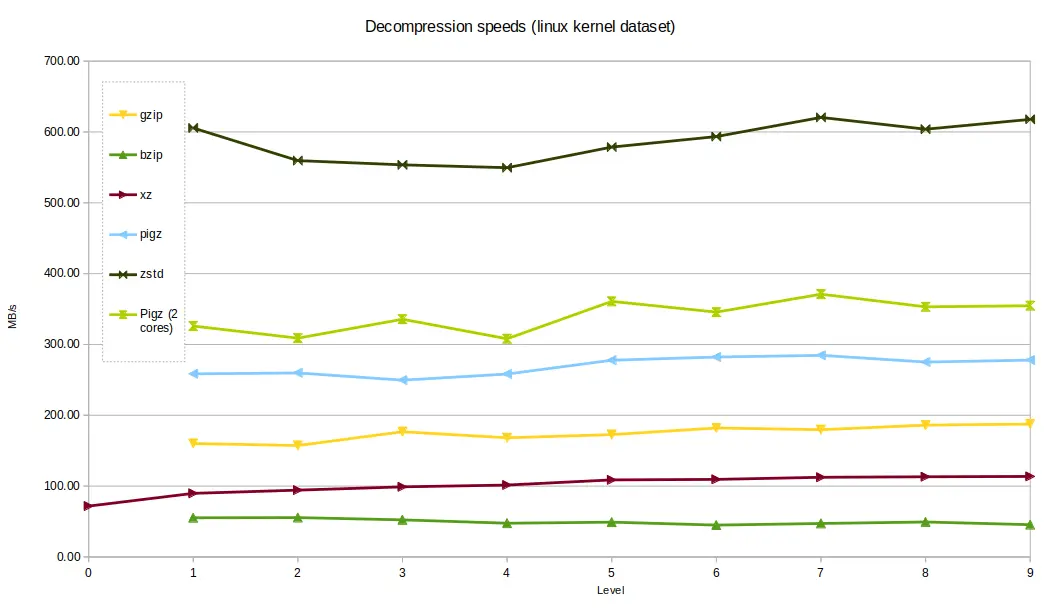
The Linux kernel dataset compressed at a higher rate than
the genomics data. zstd out-performed the other tools again, taking around 1.4
seconds to decompress, with xzand bzip2 the slowest, taking around 8
seconds and 17 seconds respectively - still not bad for a 800MB uncompressed
file.
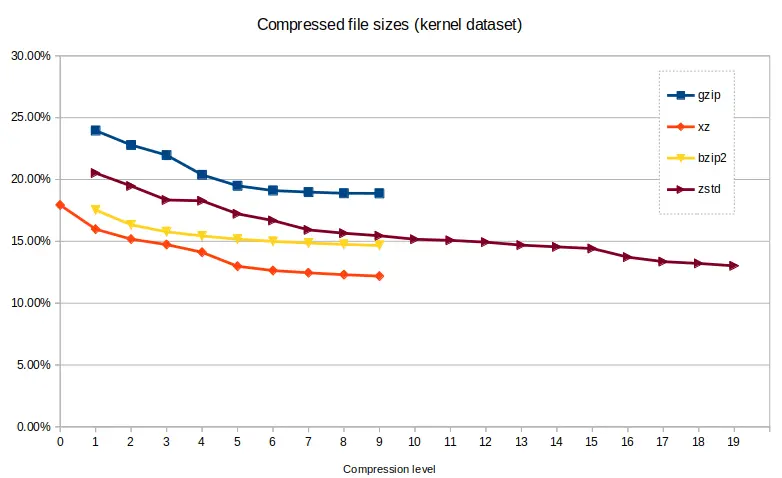
Compressed file size¶
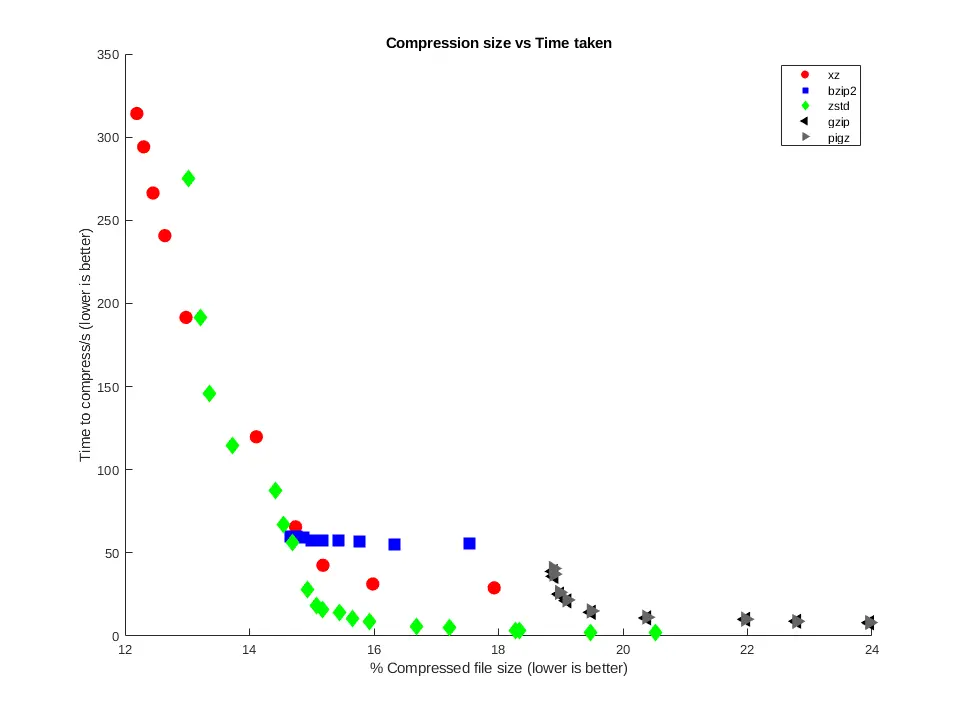
The kernel dataset compressed more effectively than the genome dataset, due to
its more varied content. xz produced the smallest compressed files with an
excellent 12.19% of the original file size, taking 314 seconds, with zstd
getting close at the higher levels, producing a 13.02% result in 275 seconds
with single-core confinement.
Comparing size/time showed the versatility of zstd across the compression
levels, able to provide fast, and high levels of compression, and sometimes
both at once. xz reached the parts that others can't reach in terms of
compression size, but with a trade-off against time. bzip2 was not
particularly versatile, operating in a narrow time/size range. gzip, while
quite fast, couldn't produce small enough file sizes compared to other tools.
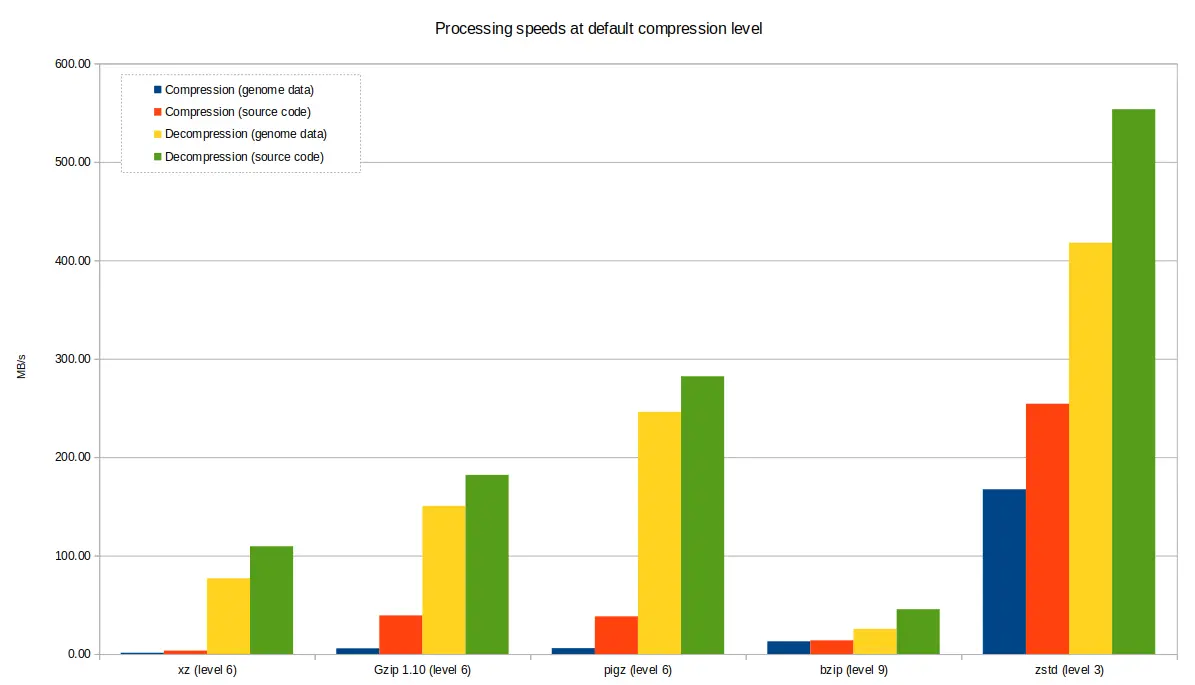
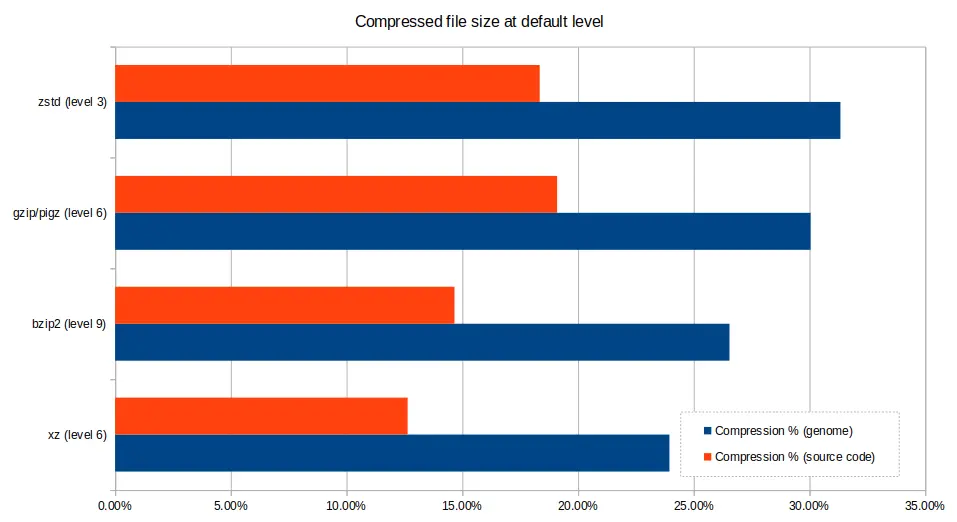
Default compression levels¶
Compression tools offer quite varied results based upon the level of compression selected by the user. The default level (i.e the level chosen by the tool if not otherwise specified) is rather arbitrary and may not be best for your use case, so it's good to appreciate that other options may be more suitable - choosing a lower compression level will still give you fairly good results and very short computation time, or conversely, if you are very constrained on space, crank up the levels and wait a bit longer for your results.
The charts below demonstrate the varying performance of the tools when
selecting the default. Note that pigz, while based on gzip, and produces
compatible .gz files of the same size, which can also be uncompressed with
gzip, decompressed considerably better. However, zstd can also be used to
compress and decompress .gz at speeds comparable to pigz, in addition to
its .zst format.
The main take-away is to choose the best level for your situation, rather than assuming the default option is best.
Summary¶
In spite of the charts showing varying performance based on the choice of tool, it's good to get some perspective. The 800MB kernel file compressed to between 12%-24% of its original size, in times ranging from 2-314 seconds with single-core compression. Each compression tool had a range of only around 5% file size difference between its slowest and fastest compression level.
For sites offering files for download, where bandwidth and storage are provided
at cost, xz format produces small file sizes, although xz was one of the
slower formats for decompression.
For large files that are accessed often, fast decompression may be be your driver.
zstd is an incredibly versatile tool, and performed very well in terms of
performance - the best all-rounder, it coped well with source code, and tricky
genome data. In addition to producing .zstd files, with use of the --format
option, it can operate on .gz and .xz files too (and lz4 format, , with
comparable performance to pigz and xz, enabling you to use a single tool to
handle most of the compressed files you encounter.
If you are already compressing your larger data files as part of your workflow,
well done. If you having traditionally been using bzip2 or gzip, maybe
consider whether you will get better performance from another tool? If you
aren't currently compressing your larger files, maybe you could consider it -
hopefully we've persuaded you that it's not as laborious as you thought. Since
Research Projects pay for their Research Data Storage, it's worth considering -
it also has a knock-on effect - it reduces time to copy files over the network,
and reduces the number of tapes we need for backups.
Title image: v2osk on Unsplash
Human Genome image: dollar_bin