Using R inside of Conda¶
Whilst most Apocrita users will want to use the R module or RStudio via OnDemand for R workflows, it is also possible to use R inside of Conda via Miniforge.
You may wish to do this when you want to have R packages available at the same time as other packages you can install via Conda, or when your workflow requires the use of both Python and R at the same time.
Setting up your environment and library¶
To run R inside of Conda, first you will need to create and activate a Conda environment according to our documentation. Once you are inside your activated environment, you can then install any R packages required.
When running R inside Conda, it is best to try to stick to installing any additional R packages you require using the Conda package manager:
https://docs.anaconda.com/free/working-with-conda/packages/using-r-language/
There are thousands of commonly used R packages for data science available from the Conda Forge channel, as well as many others in the Bioconda channel. You can use the search facility at anaconda.org to search for packages.
Do not use the defaults channel
Previous documentation may have led you to use the defaults channel in
your ~/.condarc file. This is actively recommended against in the
official Mamba documentation.
Instead, you should use nodefaults, which will disable the defaults
channel and use only the conda-forge channel.
Use Mamba instead of Conda
The official documentation for many Conda packages will often state that you
should use conda for commands such as conda install etc. We recommend
using mamba instead as it is much faster. See
this blog post for further
information.
CRAN packages¶
A large number of CRAN packages are available to install using Anaconda. You
will need to add r- before the regular package name. For instance, if you want
to install Seurat, you will need to use mamba install r-seurat or for
rJava, type mamba install r-rjava.
Pay attention to the output for the proposed installation versions of your
package and its dependencies. You might find that the default version of
something you are offered is too old and another channel offers a newer version.
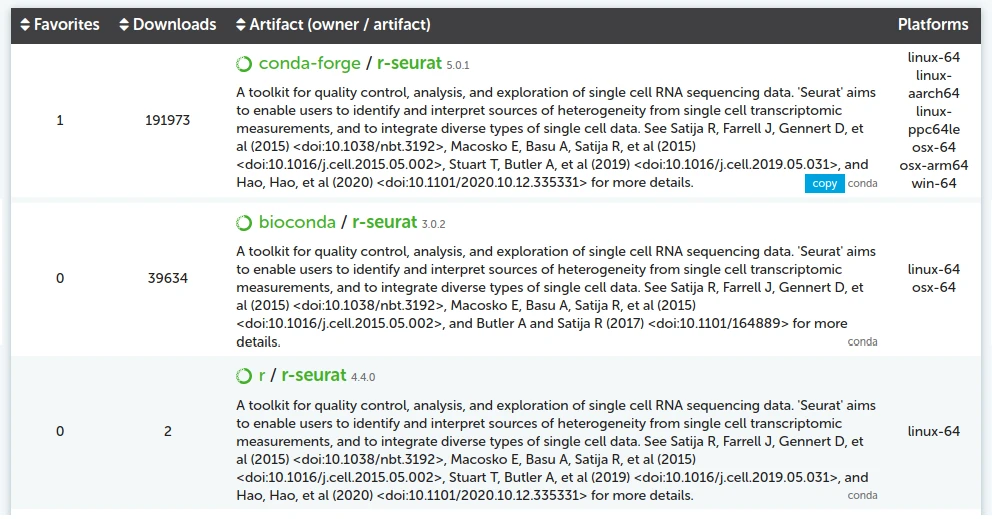
For example, if you search for the r-seurat package on anaconda.org, you
will see that it is available from multiple channels:
Miniforge will use the conda-forge channel by default. To select a specific
channel, add it to your installation command. For instance, if you wanted to
install Seurat from Bioconda:
mamba install bioconda::r-seurat
You may find that when you specify a channel in this way, then Conda will complain that some dependencies can't be fulfilled. You can specify multiple channels in your installation command, and they will be used in the order specified:
mamba install -c bioconda -c conda-forge <package name>
The above command would install your package from the Bioconda channel, and if any required dependencies aren't found in Bioconda, then the installation process will search Conda Forge for them as well.
Bioconductor packages¶
There are also a lot of Bioconductor packages
available to install using Anaconda. Most of these are available from the
Bioconda channel; you will usually need to add
bioconductor- before the regular package name. For example, to install
HIBAG:
mamba install bioconda::bioconductor-hibag
Again, depending on what packages are already in your library, you may find you need to specify additional channel(s) to fulfil all required dependencies:
mamba install -c bioconda -c conda-forge bioconductor-hibag
This will install HIBAG from the
Bioconda channel, and use
Conda Forge to fulfil any missing dependencies that aren't available from
Bioconda.
Beware install.packages¶
Whilst it may be tempting to install packages within the R shell running inside
Conda using install.packages as you might do when running R using our module
or RStudio OnDemand, it is best to try to avoid this. This might install those
packages into your personal R library (default path
~/R/x86_64-pc-linux-gnu-library/<R version>). This path may already contain
packages that have been compiled and installed using the R module or RStudio.
The issue here is that the compilation environment inside your Conda environment is likely to be markedly different from that of the R module or RStudio, particularly if you have loaded additional modules and compiled and installed packages in your personal library. There will be different versions of key packages and libraries such as GCC, and all sorts of issues can arise.
It's best wherever possible to only install R packages in your Conda environment
using the mamba install methods detailed above, so that they are compiled
consistently using the same environment, and don't get mixed up with any
packages in your personal R library. Also, using install.packages inside Conda
runs the risk that some packages might be compiled using a mix of dependencies
from both inside and outside of the Conda environment, which isn't ideal. Using
mamba install should also be markedly faster than install.packages as well.
If something is missing from all Conda channels, then you can proceed to use
installation methods such as install.packages. Just be sure to check your
personal library path carefully first:
> .libPaths()
[1] "/path/to/conda/env/lib/R/library"
This path should point to a directory inside your Conda environment. If it doesn't, then make sure that it does before installing any additional packages by running the following:
> .libPaths("/path/to/conda/env/lib/R/library")
An additional check is to run library(); the output should start with
something similar to:
Packages in library ‘/path/to/conda/env/lib/R/library’:
The only path listed should be inside your Conda environment; if there are any other library paths listed then tread very carefully.